ClinicalML - A medically interpretable ML pipeline for clinical outcome prediction using MIMIC-III discharge summaries
ClinicalML is an interpretable machine learning pipeline for predicting ICU patient outcomes—mortality and length of stay—using admission-time clinical notes. It extracts disease and drug entities, reduces dimensionality with BioClinicalBERT embeddings, and applies traditional ML models. ClinicalML performs comparably to BERT-based models while offering greater transparency and clinical trust.
Code releasing soon after clearance from PhysioNet.org
Project Summary
1. Introduction
ClinicalML is a lightweight, interpretable pipeline designed to predict clinical outcomes such as in-hospital mortality and length of stay (LoS) using textual EHR data at ICU admission. While modern language models like BERT achieve high performance, their lack of transparency hinders trust in clinical settings. Our pipeline offers comparable performance using traditional ML models, providing explainable insights to clinicians. ClinicalML achieved macro-F1 scores of 0.58 for mortality prediction and 0.33 for LoS prediction.

Overview of ClincialML pipeline. We first construct the admission notes cohort based on BioClincialBERT work. We then utilize Med7 NER tagger and HunFlair to extract drug and disease names respectively. For dimensionality reduction, we adopt K-means clustering on BioClincicalBERT embeddings of the extracted entites, to derive 256-sized clusters which are then reverse mapped on the entities to create OneHot Vectors. Using reduced OneHot feature vectors of therapeutics and diseases, we train a suite of ML models on the Mortality Prediction and Length of Stay Task.
2. Related Work
We review scoring systems (e.g., APACHE, SAPS), vital-sign based methods, and BERT-driven textual prediction. Prior models using vital signs often falter when real-time vitals are missing. Others leverage BERT but are black-box in nature. ClinicalML targets this gap by using admission-note-derived features with interpretable ML methods.
3. Dataset
We used MIMIC-III v1.4 and extracted admission-time notes from discharge summaries using Van Aken et al.'s method. Only sections available at admission (e.g., Present Illness, Medications, Family History) were included. Mortality and LoS labels were derived from ADMISSIONS.csv while avoiding data leakage. We retained 48,745 samples for mortality prediction and 43,609 for LoS.

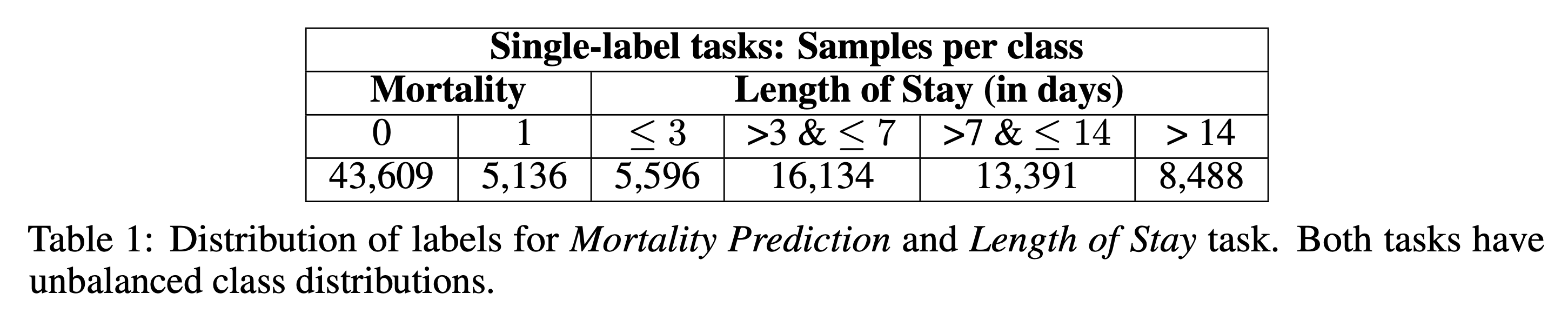
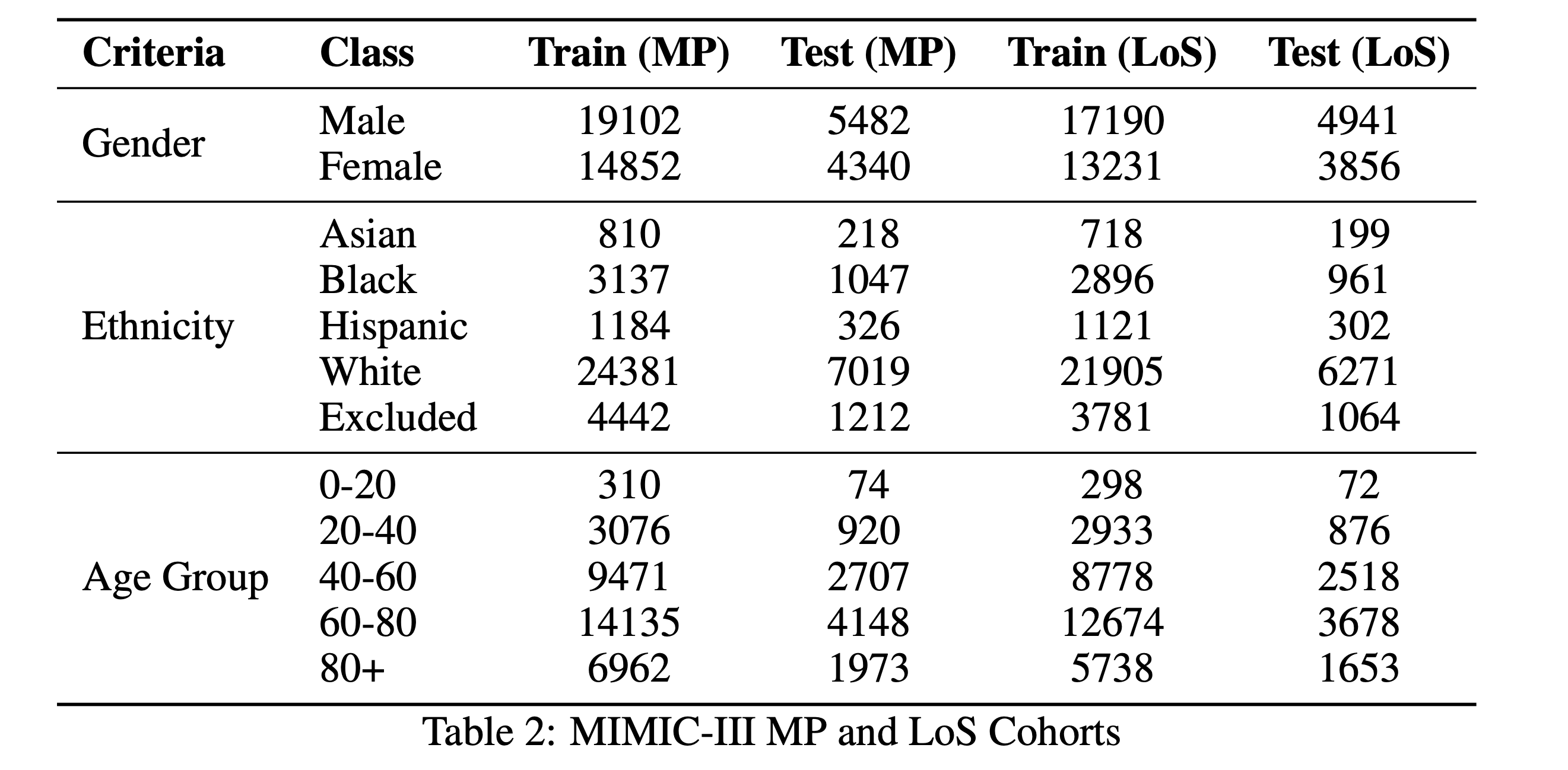
4. Methods
4.1 Feature Extraction and Selection
Our primary feature variables for predicting clinical outcome is prior chronic disease history and drugs/therapeutics prescribed to a patient during treatment as recorded in discharge summaries. The idea is to rely on minimal yet meaningful features to predict risk of mortality and elongated length of ICU stay. We applied Med7 and HunFlair NER taggers to extract therapeutics and disease mentions. BioClinicalBERT embeddings were used to encode these terms, and KMeans (via FAISS) was employed to cluster them into 256 groups. This reduced noise and redundancy.

4.2 Feature Representation
Entities were one-hot encoded based on their cluster assignments, representing disease and drug presence. Absence of features is also meaningful for predictions.
4.3 Training Classifiers
We trained four traditional machine learning classifiers—Logistic Regression, Random Forest, Gradient Boosted Trees, and XGBoost—on the one-hot encoded representations derived from clustered disease and therapeutic entities. Each model was evaluated separately for the mortality prediction and length of stay (LoS) tasks. To ensure optimal performance, we employed exhaustive hyperparameter tuning using grid search with cross-validation for each classifier. The grid search space included key parameters such as tree depth, number of estimators, and learning rate (for XGBoost and GBM). All experiments were conducted on the NYU Greene High-Performance Computing cluster via the OnDemand interface, ensuring computational efficiency and reproducibility. This rigorous training setup allowed us to identify the best-performing configurations for each task and feature set.
</p>
5. Experiments
5.1 Resampling Techniques
We compared three setups to handle class imbalance: no resampling, SMOTE, and simple oversampling. Across all tasks and features, oversampling yielded the best performance.
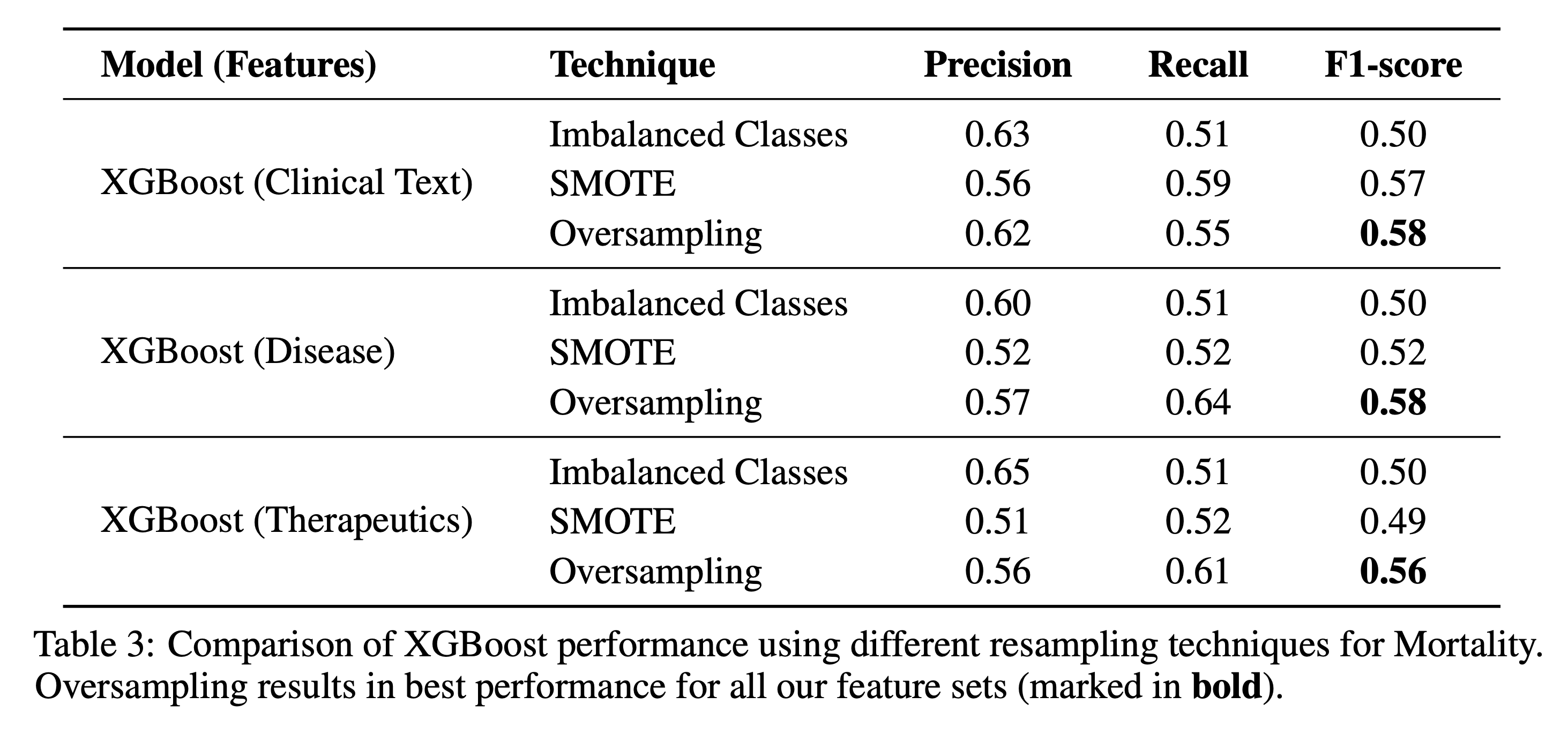
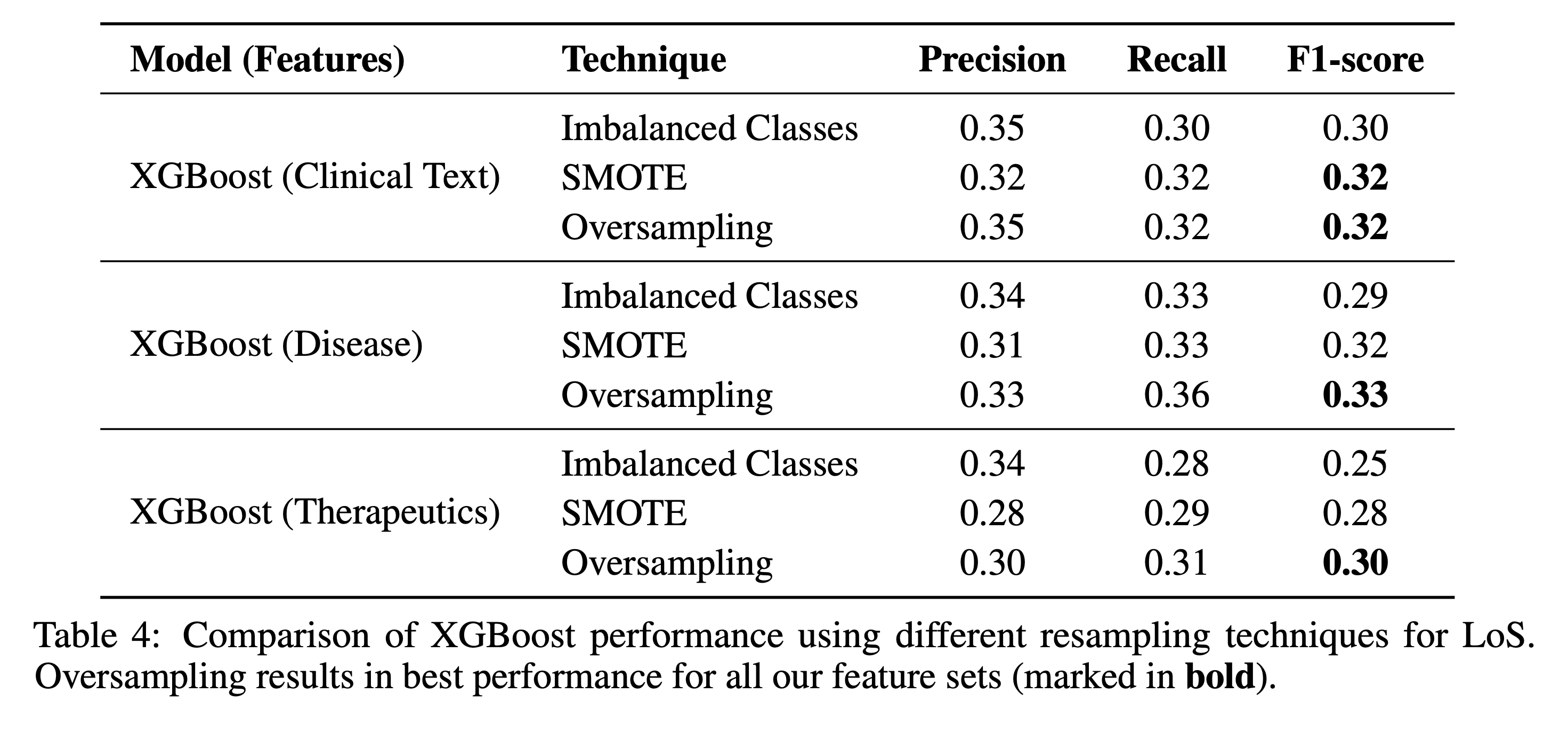
5.2 Feature Ablation Studies
- Only Therapeutics: F1 = 0.56 (MP), 0.30 (LoS)
- Only Diseases: F1 = 0.58 (MP), 0.33 (LoS)
- Doc2Vec (BioWordVec): F1 = 0.58 (MP), 0.32 (LoS)
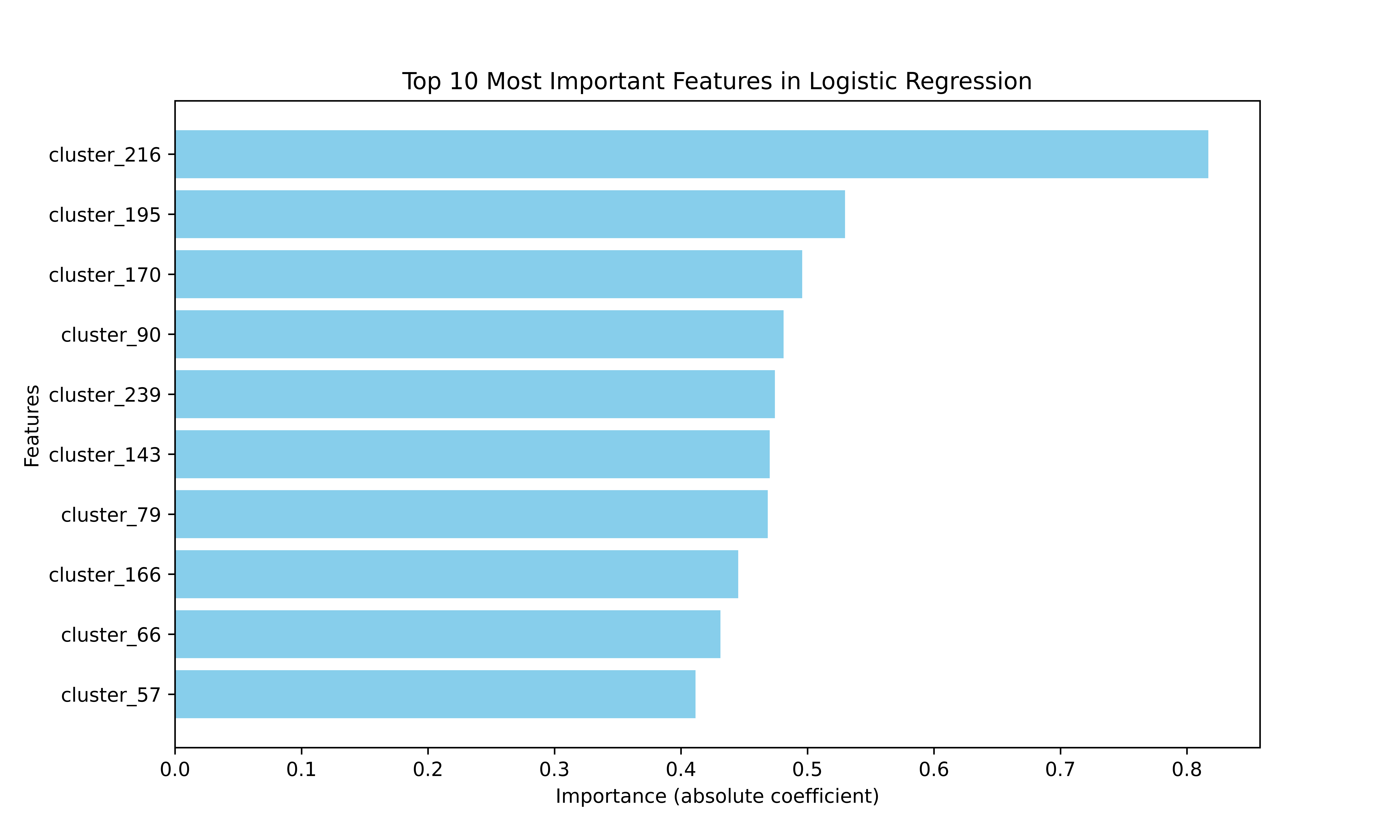
Disease features were the strongest predictors. Therapeutics showed weak correlation with mortality.

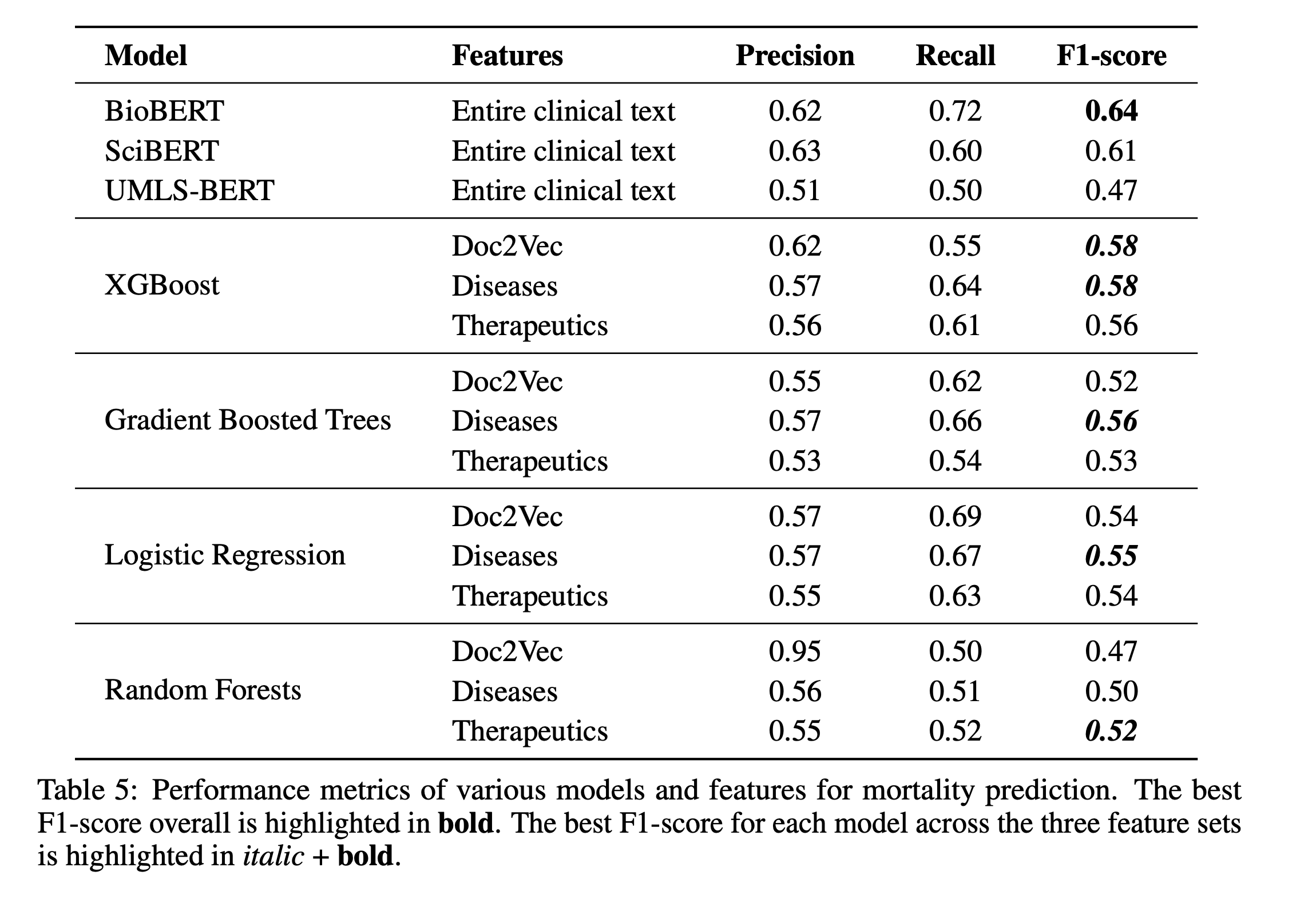
5.3 Benchmarking Against BERT Models
We compared ClinicalML against BioBERT, SciBERT, and UMLS-BERT. Though slightly underperforming, ClinicalML offered better interpretability.
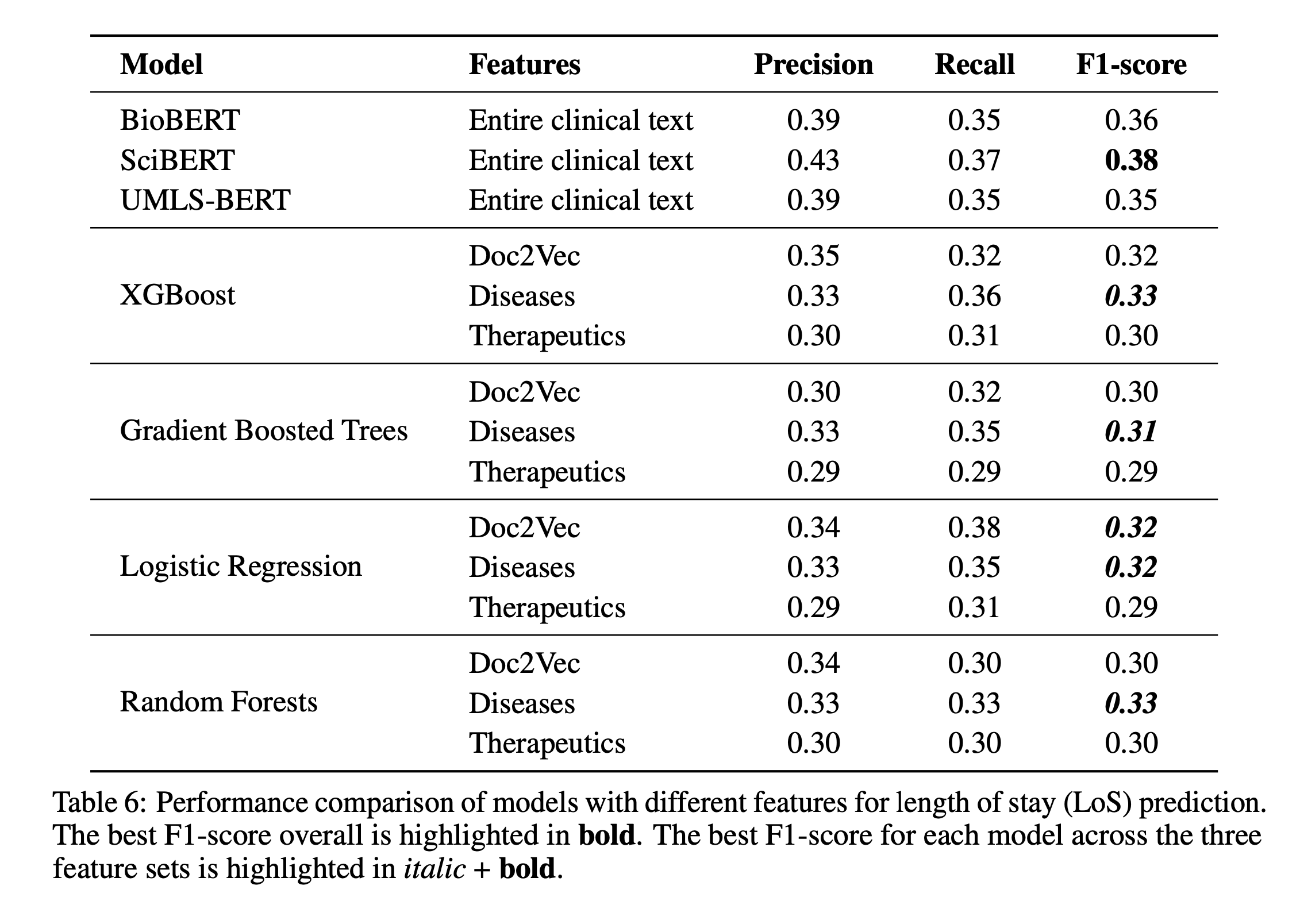
6. Discussion and Conclusion
ClinicalML bridges the gap between interpretability and performance in clinical ML tasks. Traditional ML models with engineered features from textual admission notes perform close to BERT models. Disease clusters were particularly predictive. Interpretability via feature importance builds trust in predictions. Future work includes integrating real-time temporal data and extending to multimodal settings.